Automated detection of third molars and mandibular nerve by deep learning
-
Shankeeth Vinayahalingam, Tong Xi, Stefaan Bergé, Thomas Maal & Guido de Jong
Scientific Reports (2019)
본 논문은 2019년에 발표된 논문으로, 사랑니(third molar)과 하악관 신경(mandibular nerve)를 detection하는 딥러닝 기법을 소개합니다.
논문이 어렵지는 않아서 한글 해석은 정리하지 않았고, 보면서 저에게 필요한 문단만 정리하였습니다.
출처 : www.nature.com/articles/s41598-019-45487-3#Sec2
Abstract
The approximity of the inferior alveolar nerve (IAN) to the roots of lower third molars (M3) is a risk factor for the occurrence of nerve damage and subsequent sensory disturbances of the lower lip and chin following the removal of third molars.
In this study, we developed and validated an automated approach, based on deep-learning, to detect and segment the M3(lower third molars) and IAN(Inferior alveolar nerve) on OPGs(dental panoramic radiographs).
Introduction
As with other forms of surgery, the surgical removal of the lower third molars is associated with risk for complications. One of the most distressing complications following the removal of lower third molars is damage to the inferior alveolar nerve (IAN). IAN injuries cause temporary, and in certain cases, permanent neurosensory impairments in the lower lip and chin, with an incidence of 3.5% and 0.91% respectively6. Therefore, preventing damage to the IAN is of utmost importance in the daily clinical practice.
Conventional two-dimensional panoramic radiograph, the orthopantomogram (OPG), is the most commonly used imaging technique to assess third molars and their relationship to the mandibular canal.
The aim of this present study was to achieve an automated high-performance segmentation of the third molars, and the inferior alveolar nerves (IAN) on OPG images using deep-learning as a fundamental basis for an improved and more automated risk assessment of IAN injuries.
Material and Method
The OPGs were taken with patients standing upright, head supported by the 5-point head support, with the upper and lower incisors biting gently into the bite block. A total of 81 OPGs were analysed.

A. M3 and IAN detection workflow
A workflow for the M3 detection and nerve segmentation was created using image processing and deep-learning networks (Fig. 1).
Step 1 : Dataset preprocessing
-
Resize : 2048x1024 → 1024x512
-
contrast enhancement (by determining the mean and standard deviation of the grayscale pixel values of the whole dataset.)
-
Data augmentation during deep-learning training
-
In this case, the final black and white images were mirrored vertically, horizontally and both at the same time.
-
Four variants off all given images were created in the end to aid with rotation invariance.
Step 2 & 5: the overall dentition detection and rough IAN
-
U-net
-
Loss by means of the (negative) smoothed DICE score
-
a default ADAM (Kingma and Ba 2015) optimizer with a learning rate of 1*10-4 with high momentum β-values (β1 = 0.9, β2 = 0.99) and no decay
-
stopped after 32 generations have passed.

Step 3 : Lower third molar segmentation using deep-learning
-
double-input U-net
-
This starts by two layers of 32-kernel convolution steps per image. The result of these convolutions will be concatenated and used as an input for a similar U-net
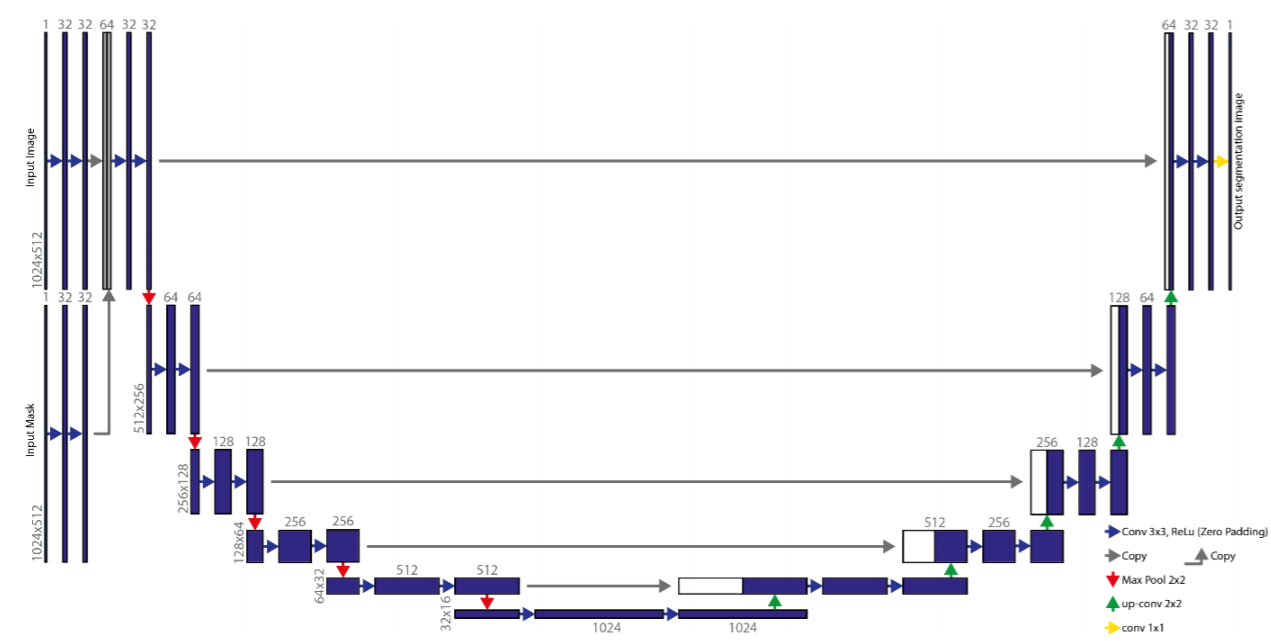
Step 4 & 6 : Cropping the M3 and IAN
-
Using the detected third molars in step 3 an automated crop of the original OPG is made in the fourth step 4. A margin extending caudally around the M3 is taken from the OPG to include a part of the IAN.


Step 7 : IAN segmentation using deep-learning
-
double u-net

B. Deep-Learning network training



Results

'ML | DL' 카테고리의 다른 글
| [Review] Real-Time High-Resolution Background Matting (0) | 2021.12.16 |
|---|---|
| Loaded runtime CuDNN library: 7.4.2 but source was compiled with 7.6.0 에러 (0) | 2021.11.17 |
| [Review] LS-Net : Fast Single-Shot Line-Segment Detector (0) | 2020.07.29 |
| [ML] Decision Tree : 의사 결정 나무 (1) (0) | 2020.07.21 |
| [Keras] fit vs fit_generator (0) | 2020.07.15 |


